Additional analyses
- Here you can find the code and the results of all additional analyses.
- To see a good overview of the results, I recommend you directly go down to the Figures section.
- But if you’re interested in how the analyses were executed, see below.
- To see the code, click on button “Code”. Alternatively, you can download the rmd file from the github repo.
Set-up
Load packages.
# install packages
# devtools::install_github("https://github.com/tdienlin/td@v.0.0.2.5")
# define packages
library(broom.mixed)
library(brms)
library(devtools)
library(GGally)
library(ggplot2)
library(gridExtra)
library(kableExtra)
library(knitr)
library(lavaan)
library(lme4)
library(magrittr)
library(mice)
library(mvnormalTest)
library(PerFit)
library(psych)
library(quanteda.textstats)
library(semTools)
library(tidyverse)
library(td)
# load workspace
load("data/workspace_2.RData")Robustness analyses
With additional covariates
These models include additional covariates that weren’t preregistered and that weren’t included, mainly because they are likely to be mediators. These are:
Trust in ORF
Trust in police
Trust in parliament
Trust in health sector
Trust in government
Trust in army
Media use: Kronen Zeitung oder www.krone.at
Media use: Der Standard oder derstandard.at
Media use: Die Presse oder diepresse.com
Media use: Oesterreich oder oe24.at
Media use: Kurier oder kurier.at
Media use: Salzburger Nachrichten oder salzburg.at
Media use: Sonstige oesterreichische Tageszeitungen
Media use: ORF (Nachrichten)
Media use: Privatfernsehen (Nachrichten)
Satisfaction with democracy
Life satisfaction
model_life_sat_lmer_add <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b +
trst_media_w + trst_police_w + trst_media_w + trst_hlthsec_w + trst_gov_w + trst_army_w +
trst_media_b + trst_police_b + trst_media_b + trst_hlthsec_b + trst_gov_b + trst_army_b
"
fit_life_sat_lmer_add <- with(d_long_100_mim_mice,
exp = lmerTest::lmer(model_life_sat_lmer_add))
fit_life_sat_lmer_add <- summary(pool(fit_life_sat_lmer_add), conf.int = TRUE)
print_res(fit_life_sat_lmer_add)Positive Affect
model_aff_pos_lmer_add <- "
aff_pos_m ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b +
trst_media_w + trst_police_w + trst_media_w + trst_hlthsec_w + trst_gov_w + trst_army_w +
trst_media_b + trst_police_b + trst_media_b + trst_hlthsec_b + trst_gov_b + trst_army_b
"
fit_aff_pos_lmer_add <- with(d_long_100_mim_mice,
exp = lmerTest::lmer(model_aff_pos_lmer_add))
fit_aff_pos_lmer_add <- summary(pool(fit_aff_pos_lmer_add), conf.int = TRUE)
print_res(fit_aff_pos_lmer_add)Negative Affect
model_aff_neg_lmer_add <- "
aff_neg_m ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b +
trst_media_w + trst_police_w + trst_media_w + trst_hlthsec_w + trst_gov_w + trst_army_w +
trst_media_b + trst_police_b + trst_media_b + trst_hlthsec_b + trst_gov_b + trst_army_b
"
fit_aff_neg_lmer_add <- with(d_long_100_mim_mice,
exp = lmerTest::lmer(model_aff_neg_lmer_add))
fit_aff_neg_lmer_add <- summary(pool(fit_aff_neg_lmer_add), conf.int = TRUE)
print_res(fit_aff_neg_lmer_add)Without control variables
Let’s inspect how results change when control variables are omitted.
Life satisfaction
model_life_sat_lmer_nco <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b
"
fit_life_sat_lmer_nco <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_life_sat_lmer_nco))
fit_life_sat_lmer_nco <- summary(pool(fit_life_sat_lmer_nco), conf.int = TRUE)
print_res(fit_life_sat_lmer_nco)Positive Affect
model_aff_pos_lmer_nco <- "
aff_pos_m ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b
"
fit_aff_pos_lmer_nco <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_pos_lmer_nco))
fit_aff_pos_lmer_nco <- summary(pool(fit_aff_pos_lmer_nco), conf.int = TRUE)
print_res(fit_aff_pos_lmer_nco)Negative Affect
model_aff_neg_lmer_nco <- "
aff_neg_m ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b
"
fit_aff_neg_lmer_nco <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_neg_lmer_nco))
fit_aff_neg_lmer_nco <- summary(pool(fit_aff_neg_lmer_nco), conf.int = TRUE)
print_res(fit_aff_neg_lmer_nco)Preregistered
Some changes were introduced because of feedback from colleagues (multiple imputation, imputation of all participants, inclusion of all variables). In what follows, please find the results as originally planned (but with some necessary deviations; for example not all preregistered variables could be included).
- Use data-set in which participants with >50% data were removed.
- Also use factor scores (possible without multiple imputation).
Note that because here no multiple imputation was used, output looks differently.
Life satisfaction
model_life_sat_lmer_pre <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_life_sat_lmer_pre <- lmerTest::lmer(model_life_sat_lmer_pre, data = d_long_50_imp)
print_res(broom.mixed::tidy(fit_life_sat_lmer_pre, conf.int = T), imputation = FALSE)Positive Affect
model_aff_pos_lmer_pre <- "
aff_pos_fs ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_pos_lmer_pre <- lmerTest::lmer(model_aff_pos_lmer_pre, data = d_long_50_imp)
print_res(broom.mixed::tidy(fit_aff_pos_lmer_pre, conf.int = T), imputation = FALSE)Negative Affect
model_aff_neg_lmer_pre <- "
aff_neg_fs ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
med_txt_kro_b + med_txt_sta_b + med_txt_pre_b + med_txt_oes_b + med_txt_kur_b + med_txt_slz_b + med_txt_son_b +
med_vid_orf_b + med_vid_pri_b +
med_txt_kro_w + med_txt_sta_w + med_txt_pre_w + med_txt_oes_w + med_txt_kur_w + med_txt_slz_w + med_txt_son_w +
med_vid_orf_w + med_vid_pri_w +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
sat_dem_w + sat_dem_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_neg_lmer_pre <- lmerTest::lmer(model_aff_neg_lmer_pre, data = d_long_50_imp)
print_res(broom.mixed::tidy(fit_aff_neg_lmer_pre, conf.int = T), imputation = FALSE)Without imputation
Final analysis, but now don’t use any imputed data whatsoever. Was preregistered as additional analysis to provide comparison.
Life satisfaction
model_life_sat_lmer_noi <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_life_sat_lmer_noi <- lmerTest::lmer(model_life_sat_lmer_noi, data = d_long_50)
print_res(broom.mixed::tidy(fit_life_sat_lmer_noi, conf.int = T), imputation = FALSE)Positive Affect
model_aff_pos_lmer_noi <- "
aff_pos_fs ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_pos_lmer_noi <- lmerTest::lmer(model_aff_pos_lmer_noi, data = d_long_50)
print_res(broom.mixed::tidy(fit_aff_pos_lmer_noi, conf.int = T), imputation = FALSE)Negative Affect
model_aff_neg_lmer_noi <- "
aff_neg_fs ~
(1 | id) + (1 | wave) +
soc_med_read_w + soc_med_like_share_w + soc_med_post_w +
soc_med_fb_w + soc_med_ig_w + soc_med_wa_w + soc_med_yt_w + soc_med_tw_w +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_neg_lmer_noi <- lmerTest::lmer(model_aff_neg_lmer_noi, data = d_long_50)
print_res(broom.mixed::tidy(fit_aff_neg_lmer_noi, conf.int = T), imputation = FALSE)Figure
- In what follows, see figure with all results combined.
- Results show that different models are comparable.
- The model without control variables stands out a bit, showing that controlling for additional varying variables makes a difference.
- The preregistered model without imputation also has larger confidence intervals, which was to be expected given fewer data.
# get data
dat_fig_results_activity_pub <- get_dat_res(
fit_aff_neg_lmer_pub, fit_aff_pos_lmer_pub, fit_life_sat_lmer_pub,
type = "Activity", analysis = "1. Final, preferred", variance = "within")
dat_fig_results_channel_pub <- get_dat_res(
fit_aff_neg_lmer_pub, fit_aff_pos_lmer_pub, fit_life_sat_lmer_pub,
type = "Channels", analysis = "1. Final, preferred", variance = "within")
dat_fig_results_activity_add <- get_dat_res(
fit_aff_neg_lmer_add, fit_aff_pos_lmer_add, fit_life_sat_lmer_add,
type = "Activity", analysis = "2. Additional covariates", variance = "within")
dat_fig_results_channel_add <- get_dat_res(
fit_aff_neg_lmer_add, fit_aff_pos_lmer_add, fit_life_sat_lmer_add,
type = "Channels", analysis = "2. Additional covariates", variance = "within")
dat_fig_results_activity_nco <- get_dat_res(
fit_aff_neg_lmer_nco, fit_aff_pos_lmer_nco, fit_life_sat_lmer_nco,
type = "Activity", analysis = "3. No covariates", variance = "within")
dat_fig_results_channel_nco <- get_dat_res(
fit_aff_neg_lmer_nco, fit_aff_pos_lmer_nco, fit_life_sat_lmer_nco,
type = "Channels", analysis = "3. No covariates", variance = "within")
dat_fig_results_activity_pre <- get_dat_res(
fit_aff_neg_lmer_pre, fit_aff_pos_lmer_pre, fit_life_sat_lmer_pre,
type = "Activity", analysis = "4. As preregistered", variance = "within")
dat_fig_results_channel_pre <- get_dat_res(
fit_aff_neg_lmer_pre, fit_aff_pos_lmer_pre, fit_life_sat_lmer_pre,
type = "Channels", analysis = "4. As preregistered", variance = "within")
dat_fig_results_activity_noi <- get_dat_res(
fit_aff_neg_lmer_noi, fit_aff_pos_lmer_noi, fit_life_sat_lmer_noi,
type = "Activity", analysis = "5. No imputation", variance = "within")
dat_fig_results_channel_noi <- get_dat_res(
fit_aff_neg_lmer_noi, fit_aff_pos_lmer_noi, fit_life_sat_lmer_noi,
type = "Channels", analysis = "5. No imputation", variance = "within")
dat_fig_results_comp <- rbind(
dat_fig_results_activity_pub,
dat_fig_results_channel_pub,
dat_fig_results_activity_nco,
dat_fig_results_channel_nco,
dat_fig_results_activity_add,
dat_fig_results_channel_add,
dat_fig_results_activity_pre,
dat_fig_results_channel_pre,
dat_fig_results_activity_noi,
dat_fig_results_channel_noi
)
# make figure
fig_results_comp <- make_graph_res(
dat_fig_results_comp,
sesoi = "est",
facet = "type"
)
fig_results_comp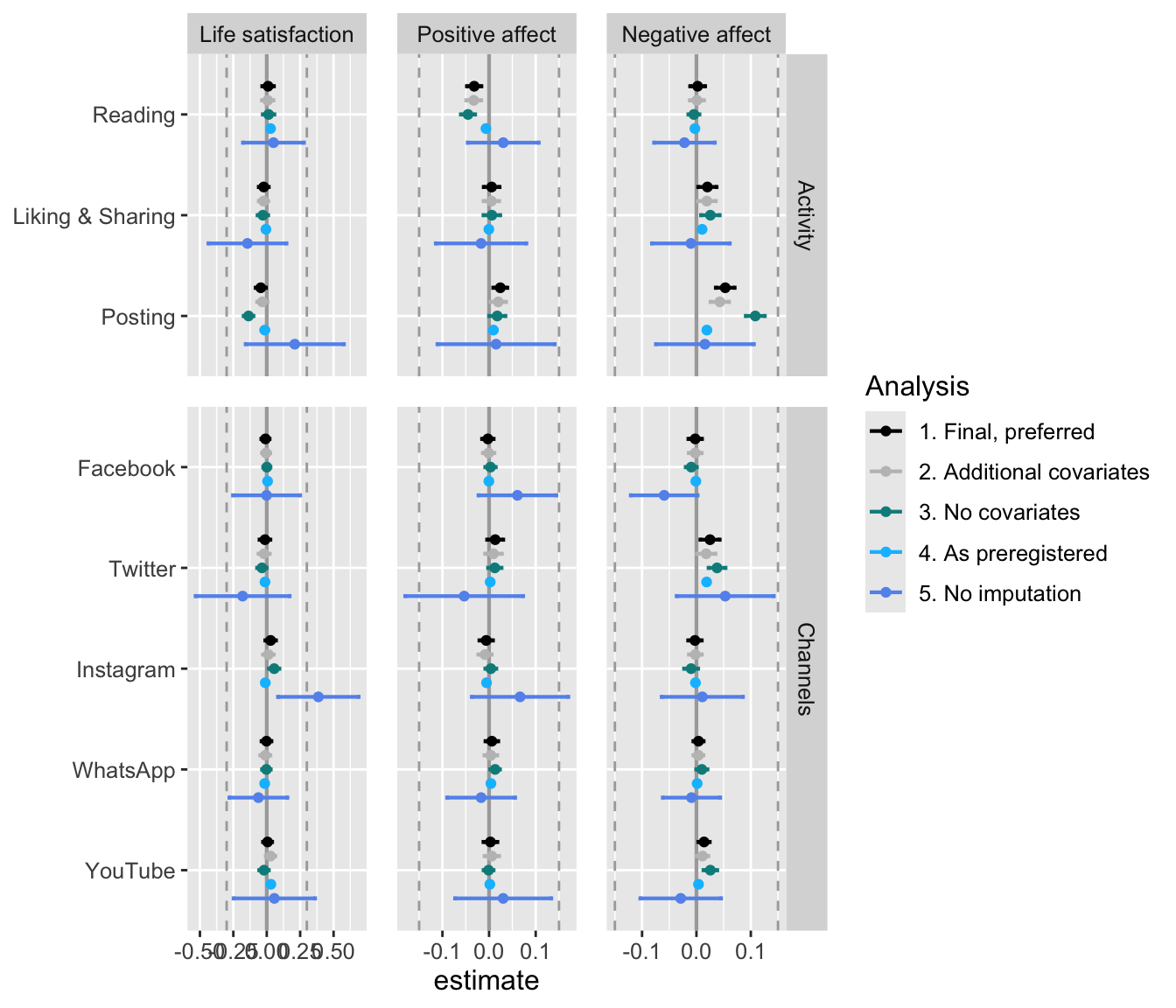
# safe figure
ggsave("figures/fig_results_comp.png",
width = 7, height = 7,
plot = fig_results_comp)Exploratory analyses
1 Month interval
Let’s now inspect effects of media use on well-being some time later (i.e., 1 month or 4 months). This is especially relevant for life-satisfaction, which is more stable.
Let’s first format data to introduce lags.
# define waves with 1 month lag to well-being
waves_predictors_1m <- c(5, 11, 18, 24, 29)
# define waves where media use was measured
waves_media_use <- c(1, 8, 17, 23, 28)
waves_include_1m <- c(waves_media_use, waves_predictors_1m)
d_long_100_mim_mice_1m <-
d_long_100_mim_mice %>%
complete(
'long',
include = TRUE
) %>%
filter(
wave %in% waves_include_1m
) %>%
group_by(id) %>%
mutate(
life_sat = lead(life_sat, n = 1, default = NA),
aff_pos_m = lead(aff_pos_m, n = 1, default = NA),
aff_neg_m = lead(aff_neg_m, n = 1, default = NA),
) %>%
ungroup() %>%
as.mids() %>%
# select waves used for analyses
filter(
wave %in% waves_predictors_1m
)
waves_predictors_4m <- c(13, 15, 21, 25, 32)
waves_include_4m <- c(waves_media_use, waves_predictors_4m)
d_long_100_mim_mice_4m <-
d_long_100_mim_mice %>%
complete(
'long',
include = TRUE
) %>%
filter(
wave %in% waves_include_4m
) %>%
group_by(id) %>%
mutate(
life_sat = lead(life_sat, n = 1, default = NA),
aff_pos_m = lead(aff_pos_m, n = 1, default = NA),
aff_neg_m = lead(aff_neg_m, n = 1, default = NA),
) %>%
ungroup() %>%
as.mids() %>%
# select waves used for analyses
filter(
wave %in% waves_predictors_4m
)Life satisfaction
fit_life_sat_lmer_1m <- with(d_long_100_mim_mice_1m, exp = lmerTest::lmer(model_life_sat_lmer_pub))
fit_life_sat_lmer_1m <- summary(pool(fit_life_sat_lmer_1m), conf.int = TRUE)
print_res(fit_life_sat_lmer_1m)We find no significant effects.
Positive Affect
fit_aff_pos_lmer_1m <- with(d_long_100_mim_mice_1m, exp = lmerTest::lmer(model_aff_pos_lmer_pub))
fit_aff_pos_lmer_1m <- summary(pool(fit_aff_pos_lmer_1m), conf.int = TRUE)
print_res(fit_aff_pos_lmer_1m)We find no significant effects.
Negative Affect
fit_aff_neg_lmer_1m <- with(d_long_100_mim_mice_1m, exp = lmerTest::lmer(model_aff_neg_lmer_pub))
fit_aff_neg_lmer_1m <- summary(pool(fit_aff_neg_lmer_1m), conf.int = TRUE)
print_res(fit_aff_neg_lmer_1m)We find no significant effects.
4 Months Interval
Let’s not see if we find effects looking at longer intervals.
Life satisfaction
fit_life_sat_lmer_4m <- with(d_long_100_mim_mice_4m, exp = lmerTest::lmer(model_life_sat_lmer_pub))
fit_life_sat_lmer_4m <- summary(pool(fit_life_sat_lmer_4m), conf.int = TRUE)
print_res(fit_life_sat_lmer_4m)We find no significant effects.
Positive Affect
fit_aff_pos_lmer_4m <- with(d_long_100_mim_mice_4m, exp = lmerTest::lmer(model_aff_pos_lmer_pub))
fit_aff_pos_lmer_4m <- summary(pool(fit_aff_pos_lmer_4m), conf.int = TRUE)
print_res(fit_aff_pos_lmer_4m)We find no significant effects.
Negative Affect
fit_aff_neg_lmer_4m <- with(d_long_100_mim_mice_4m, exp = lmerTest::lmer(model_aff_neg_lmer_pub))
fit_aff_neg_lmer_4m <- summary(pool(fit_aff_neg_lmer_4m), conf.int = TRUE)
print_res(fit_aff_neg_lmer_4m)We find no significant effects.
Gender
Let’s next see if effects differ for males and females.
Life satisfaction
model_life_sat_lmer_male <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w * male + soc_med_like_share_w * male + soc_med_post_w * male +
soc_med_fb_w * male + soc_med_ig_w * male + soc_med_wa_w * male + soc_med_yt_w * male + soc_med_tw_w * male +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_life_sat_lmer_male <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_life_sat_lmer_male))
fit_life_sat_lmer_male <- summary(pool(fit_life_sat_lmer_male), conf.int = TRUE)
print_res(fit_life_sat_lmer_male)Interaction effects are insignificant, showing that effects don’t differ across genders.
Positive Affect
model_aff_pos_lmer_male <- "
aff_pos_m ~
(1 | id) + (1 | wave) +
soc_med_read_w * male + soc_med_like_share_w * male + soc_med_post_w * male +
soc_med_fb_w * male + soc_med_ig_w * male + soc_med_wa_w * male + soc_med_yt_w * male + soc_med_tw_w * male +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_pos_lmer_male <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_pos_lmer_male))
fit_aff_pos_lmer_male <- summary(pool(fit_aff_pos_lmer_male), conf.int = TRUE)
print_res(fit_aff_pos_lmer_male)Interaction effects are insignificant, showing that effects don’t differ across genders.
Negative Affect
model_aff_neg_lmer_male <- "
aff_neg_m ~
(1 | id) + (1 | wave) +
soc_med_read_w * male + soc_med_like_share_w * male + soc_med_post_w * male +
soc_med_fb_w * male + soc_med_ig_w * male + soc_med_wa_w * male + soc_med_yt_w * male + soc_med_tw_w * male +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
age + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_neg_lmer_male <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_neg_lmer_male))
fit_aff_neg_lmer_male <- summary(pool(fit_aff_neg_lmer_male), conf.int = TRUE)
print_res(fit_aff_neg_lmer_male)Interaction effects are insignificant, showing that effects don’t differ across genders.
Age
Let’s next inspect if effects differ across age. We’ll look at generations (Gen Z, Gen Y, …). Let’s first transform data to make age categories.
d_long_100_mim_mice <-
d_long_100_mim_mice %>%
complete(
'long',
include = TRUE
) %>%
mutate(
age_gen = ifelse(
age > 2022 - 1946,
"Silent",
ifelse(
age > 2022 - 1965,
"Boomer",
ifelse(
age > 2022 - 1981,
"Gen X",
ifelse(
age > 2022 - 1997,
"Gen Y",
"Gen Z"
)
)
)
)
) %>%
mutate(
age_gen = factor(
age_gen,
c("Gen X", "Gen Z", "Gen Y", "Boomer", "Silent")
)
) %>%
as.mids()
d_long_100_mim_mice %>%
complete(
'long',
include = TRUE
) %>%
select(age_gen) %>%
table()## age_gen
## Gen X Gen Z Gen Y Boomer Silent
## 595476 721854 763980 469812 48552Life satisfaction
model_life_sat_lmer_age <- "
life_sat ~
(1 | id) + (1 | wave) +
soc_med_read_w * age_gen + soc_med_like_share_w * age_gen + soc_med_post_w * age_gen +
soc_med_fb_w * age_gen + soc_med_ig_w * age_gen + soc_med_wa_w * age_gen + soc_med_yt_w * age_gen + soc_med_tw_w * age_gen +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_life_sat_lmer_age <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_life_sat_lmer_age))
fit_life_sat_lmer_age <- summary(pool(fit_life_sat_lmer_age), conf.int = TRUE)
print_res(fit_life_sat_lmer_age) %>%
filter(grepl(".*:.*", term))Interaction effects are insignificant, showing that effects don’t differ across generations.
Positive Affect
model_aff_pos_lmer_age <- "
aff_pos_m ~
(1 | id) + (1 | wave) +
soc_med_read_w * age_gen + soc_med_like_share_w * age_gen + soc_med_post_w * age_gen +
soc_med_fb_w * age_gen + soc_med_ig_w * age_gen + soc_med_wa_w * age_gen + soc_med_yt_w * age_gen + soc_med_tw_w * age_gen +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_pos_lmer_age <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_pos_lmer_age))
fit_aff_pos_lmer_age <- summary(pool(fit_aff_pos_lmer_age), conf.int = TRUE)
print_res(fit_aff_pos_lmer_age) %>%
filter(grepl(".*:.*", term))Interaction effects are insignificant, showing that effects don’t differ across generations.
Negative Affect
model_aff_neg_lmer_age <- "
aff_neg_m ~
(1 | id) + (1 | wave) +
soc_med_read_w * age_gen + soc_med_like_share_w * age_gen + soc_med_post_w * age_gen +
soc_med_fb_w * age_gen + soc_med_ig_w * age_gen + soc_med_wa_w * age_gen + soc_med_yt_w * age_gen + soc_med_tw_w * age_gen +
soc_med_read_b + soc_med_like_share_b + soc_med_post_b +
soc_med_fb_b + soc_med_ig_b + soc_med_wa_b + soc_med_yt_b + soc_med_tw_b +
male + born_aus + born_aus_prnts + edu_fac + employment_fac +
res_vienna + acc_bal + acc_gar + home_sqm +
corona_pos_b + corona_pos_w +
work_h_b + work_h_w +
work_homeoff_b + work_homeoff_w +
hh_income_b + hh_income_w +
hh_adults + hh_child18 + hh_child17 + hh_child14 + hh_child5 + hh_child2 +
hh_oldfam + hh_outfam + hh_partner +
home_owner +
risk_prop_b + risk_prop_w +
act_wrk_w + act_spo_w + act_frn_w + act_sho_w + act_pet_w +
act_wrk_b + act_spo_b + act_frn_b + act_sho_b + act_pet_b +
health_w + health_b +
loc_cntrl_int_m_w + loc_cntrl_int_m_b
"
fit_aff_neg_lmer_age <- with(d_long_100_mim_mice, exp = lmerTest::lmer(model_aff_neg_lmer_age))
fit_aff_neg_lmer_age <- summary(pool(fit_aff_neg_lmer_age), conf.int = TRUE)
fit_aff_neg_lmer_age_tab <- print_res(fit_aff_neg_lmer_age)fit_aff_neg_lmer_age_tab %>%
filter(grepl(".*:.*", term))Interaction effects are insignificant, showing that effects don’t differ across generations. However, there’s one exception: Gen Z differs significantly from Gen X in terms of negative affect experienced when posting more than usual COVID-19 related content.
save.image("data/workspace_3.RData")